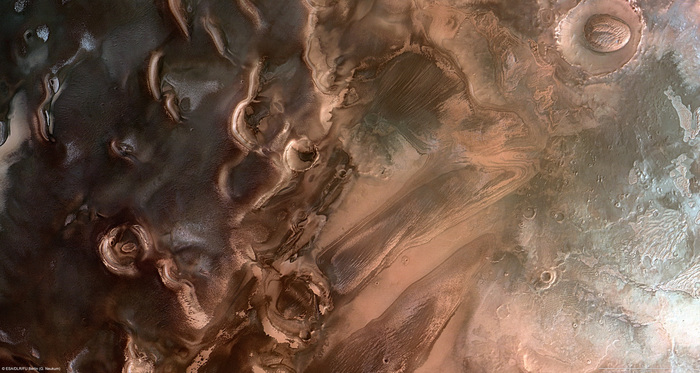
Kap København is so far north of Greenland that it faces directly towards the North Pole.
Today it is what they call a polar desert, with a few days a year free of ice that take advantage of lichens and moss to survive.
But a study has just shown that two million years ago it was almost paradise on Earth.
The authors have found evidence that hundreds of species lived there.
In a forest of birch, poplar and pine trees, aromatic and floral plants related to roses abounded.
Small herbivores, such as mice, lemmings, and rabbits, thrived on this flora.
But also big, like the current reindeer.
There were even megaherbivores, such as now extinct mastodons.
The surprising thing is that they have not found fossils of such biodiversity.
The recovery of increasingly ancient genetic material is rewriting the history of life.
DNA is a fragile material, which deteriorates when exposed to the environment for many reasons (erosion, enzyme action, temperature, pressure, oxidation...).
Where it is best preserved is in the remains of fossils that are also best preserved, such as teeth and bones.
Advances in recovery and sequencing techniques in the last two decades have allowed us to go back further and further back.
Twenty years ago, science believed that ancestral DNA that was more than 100,000 years old could not be recovered.
A decade ago, Spanish scientists managed to extract mitochondrial DNA (which is found inside the cell, but outside the cell nucleus) from humans who lived in Atapuerca 400,000 years ago.
In 2013,
managed to sequence the genome of an ancestor of horses that lived 700,000 years ago.
But last year, another group of researchers marked a milestone by extracting genetic information from mammoths preserved in the Siberian permafrost for more than a million years.
Now, the DNA found at Kap København doubles the mark, dating back to between 2.1 and 1.9 million years ago.
This time it was not a fossil.
This time they have recovered it from the ground, literally.
Before and after the ecosystem of Kap København, in the extreme north of Greenland, with a reconstruction of two million years ago.
On the right, current landscape of the now arctic desert.
Professor Svend Funder, pictured, discovered the site more than 40 years ago and was always convinced that the "North Pole Forest" was there.
Beth Zaiken / Kurt H. Kjær
The work, published in the scientific journal
Nature
, is led by Eske Willerslev, a researcher at both the University of Cambridge (United Kingdom) and director of the Center for GeoGenetics at the University of Copenhagen (Denmark).
“This is the longest story I've ever been involved in, because we started in 2006 when we went to Greenland to collect samples,” he said in an online presentation to several journalists.
With the traditional techniques of paleontology, in the coastal wasteland of Kap København they had barely found a few dozen pollen grains from plants from the past and a single fossil, that of what they believe was a hare or a rabbit.
So there were no fossils from which to extract DNA.
Even so, they were accumulating layers of earth with the hope of extracting some information from them as the technique advanced.
“Every time there were improvements in issues of extraction [of genetic material] or in sequencing technologies, we went back to review the samples, but we failed, we failed, and we failed again,” he adds.
But everything changed two years ago.
One of the new things we explored here was looking at how DNA binds to minerals.
Karina Sand, researcher at the Center for GeoGenetics at the University of Copenhagen
For barely five years, various scientific groups have been studying the possibility of extracting genetic information not from the organism to which it belonged, but from the environment in which it was found.
The brand new discipline focuses on environmental DNA, specifically ancient DNA in sediments (sedaDNA, for its acronym in English).
In one of these groups was Karina Sand, now also at the University of Copenhagen and a co-author of the study.
“One of the new things we explored here was to see how DNA binds to minerals,” she says.
Once the living being to which it belonged has decomposed, the genetic material adheres to the substrate.
Thus, they have verified that some sediments absorb more DNA than others.
“Clay minerals can preserve DNA much better than quartz minerals, for example,” explains Ella Sand.
One of the problems they found is that the clay absorbs it so well that it costs a lot to remove it later, up to four times more than quartz.
Finding genetic material in the geological substrate is already a breakthrough, but managing to extract and analyze it is what deserved to be published in Nature.
The Spanish scientist Antonio Fernández Guerra, also from the Danish Geogenetics Center and co-author of the research, recounts in a telephone conversation what it was like to work with the environmental DNA discovered in Kap København: “You have to think about what it was like two million years ago.
You had the type of forest that we described, with the trees, the plants, the streams that carried all kinds of material to the sea.
At the mouth, DNA accumulates, especially from plants.
With animals it is more difficult.
And any organism that was living on the coast two million years ago would also be in the environmental DNA.
When you take a sample of genetic sequences, there you have a bunch of bacteria and archaea,
so you have to go through all that noise to try to find that DNA that was there waiting to be found for so long.
There are times that for every million sequences that we recover, only one is valid”.
So, counting needles among haystacks, the researchers identified more than a hundred genera of vegetable plants.
The genus is one of the categories in which life is organized (taxonomic categories are the domain, the kingdom, the phylum, the class, the order, the family, the genus and, finally, the species).
Identifying specific species in environmental DNA from two million years ago shouldn't be easy, but they've done it.
Below the genus, they have identified specific trees, such as arctic willow or dwarf birch.
With animals they had it more complicated.
As in any ecosystem, there are fewer animals than plants, so they identified less animal DNA.
They did not manage to go down to the family or genus level, although they have found genetic remains of vertebrates still extant in Greenland, such as a Nearctic lemming or the arctic hare,
the only animal of which fossil remains had been found.
They have also identified genera of fleas, ants, and extinct relatives of elephants and reindeer today.
What they have not found have been carnivores.
It has its logic, following the pyramidal structure of life, the higher on the scale, the lower the number.
The researchers are convinced that, as in other ecosystems in arctic latitudes, there must have been wolves, bears and even saber-tooth tigers.
Willerslev, the senior author of this research, tells it: “Obviously, plants are more common than herbivores and herbivores, more common than carnivores.
That is probably the reason we have not found carnivores.
However, I would say that if we continue to sequence, take samples and continue to sequence, my prediction is that at some point we would capture some of the carnivores."
The fact that we are able to detect the presence of species in ecosystems for which we have no fossil evidence is fascinating.
David Díez del Molino, researcher at the Center for Paleogenetics in Stockholm
David Díez del Molino is a researcher at the Center for Paleogenetics in Stockholm (Sweden) and one of those who recovered the DNA of those mammoths from a million years ago.
Not related to the current study, he highlights the novelty of his techniques and the fruits they could bear.
“Ancient DNA from sediments is one of the hottest topics and one of the hottest fields in paleogenetics right now,” he says.
“It can allow us to reconstruct the DNA of an entire ecosystem from the past at a given site, something that with DNA from bones and other materials is basically impossible,” he adds.
But his strength is also his weakness: “The amount of DNA recovered from each species is quite small compared to DNA from bones.
For that reason, most of the analyzes that it allows for each species are a bit rudimentary ”, he completes.
In the case of their mammoth molars, for example, they were able to recover millions of nuclear sequences from three samples, allowing them to perform complex phylogenetic analyzes and discover previously unknown mammoth lineages.
But he continues to consider "fascinating the fact that we are able to detect the presence of species in ecosystems for which we have no fossil evidence and, therefore, were unknown in that place."
You can follow MATERIA on
,
and
, or sign up here to receive
our weekly newsletter
.

/cloudfront-eu-central-1.images.arcpublishing.com/prisa/NDRRW5VLERCZBFAVKVT7UBM2KE.jpg)